|
Introduction: Neural Motor Prosthetics
Decades and decades of work have gone into understanding how the motor cortex - the part of the brain that controls movement - works. Thousands of researchers have electrically recorded from motor cortex in various species (including humans) performing various motor tasks, to try to get a handle on how the motor cortex says "move my hand over" or "shake my head". Recently, our understanding of this area has gotten so good that we can start to "translate" (decode) the language of motor cortex... We'd like to say: "oh, this particular set of cells is active right now, that means this brain must be trying to move its foot to the left". What can we do with that knowledge? I worked for some time with the Donoghue Lab in the Neuroscience Department at Brown University, on neural decoding for neural prosthetic devices, and I am currently working with CyberKinetics, a medical device company aiming to bring this technology to clinical applications. The general goal of this work is to allow quadriplegic, locked-in, or otherwise motor-impaired patients to interact with their environment, through neural recording electrodes placed in the motor cortex. In the long-term vision, a patient who has lost function in his arms and legs might be able to control a robot, wheelchair, or computer desktop just by thinking about moving his or her arms. |
|
Initial Results
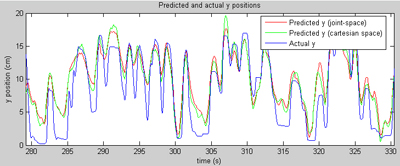
This page demonstrates some preliminary results that I generated on various preclinical (non-human) datasets collected at Brown University. The general approach is to collect data about how a subject's arm is moving (we do that at Brown using a digitizing tablet from Wacom) (a joystick-like device), while simultaneously collecting neural data from an electrical recording array. The recording array is implanted in the primary motor cortex, the part of the brain most directly responsible for controlling voluntary muscle movement. In a perfect world, we should be able to learn the mapping from neural signals to arm movements, and predict where the arm is based on the recorded neural information. So the results on this page are mostly movies that show a known arm position (we know where the arm was because we were recording the position of the tablet) and a predicted arm position (based on neural data). If we're doing our job perfectly, the two points should line up all the time. In reality, it's impossible to exactly predict arm position for a variety of reasons. Most importantly, we're only recording a small fraction of the neurons that actually control the arm, so it's like trying to piece together a novel given just the first word of each sentence. That's a problem that can only be fixed with new recording technology. Also, we still don't know exactly what the right way to decode neural signals is... and that's the problem that is an area of active research in several labs across the country. Generally about two minutes of training data was used for each of these examples, and the predictions are generated on another chunk of data collected on the same day. All of the results generated here were based on spikes alone, although the recording hardware used provides field potential data also (another topic of further investigation). I should point out that the results presented here are not inherently novel; they are based on my implementations of the work of Warland et al and Wu et al. Both of those papers are excellent references on the numerical basis of continuous neural decoding.
|
|
Example Matlab Code
I provide some sample Matlab code here. This is a simple implementation of the linear and kalman filters, along with some synthetic data and the background paper describing each filter. My implementation of joint-space decoding - described below - is included as a simple example of something you might do to build on the classic decoding algorithms. |
|
TG2: A Novel Experimental Platform (my work at Brown)
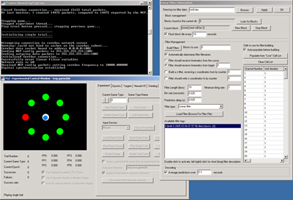
My primary contribution to the work at Brown is a software suite - TG2 - that allows experimenters to generate complex experiments using a graphical interface. The software provides a variety of experimental logic, graphic rendering, hardware interfacing, neural data processing, and data logging features. Neuroscience and psychophysics researchers can access these features graphically in their experiments, without being fluent in C++ or other low-level languages. Experimenters who are familiar with Matlab but who don't want to deal with the low-level details of graphic rendering, accurate data logging and timing, talking to neural interface devices, etc., can use a real-time Matlab interface to communicate with TG2. TG2 also aims to make experiments re-usable across different input devices and different neural recording systems by presenting the user with an abstract interface to these devices. This allows new devices to be swapped in without re-writing experiments to include device-specific interfaces or re-writing post-experimental analysis programs. Additionally, and perhaps most importantly, the software incorporates several continuous and discrete neural decoding algorithms, so researchers can perform higher-level decoding experiments without re-inventing the wheel in terms of decoding.
This package is currently the primary experimental platform in the
Donoghue Lab at Brown and in ongoing clinical experiments at Brown (based
on acute intraoperative recordings). It is also the basis for my
work at CyberKinetics. An overview of the features of TG2 is presented here. Some shots of TG2 and his friends (tools that ship in the TG2 package): |
|
Publications and References
The data shown here are preclinical, but Brown and CyberKinetics have begun to apply the implementations behind these results in clinical environments. Some information has recently been presented as posters at various conferences; all of the following make use of TG2:
I'm only mentioning work here with which I was involved, but of course there are numerous groups working on similar problems... a few of them include:
|